For the first time in a long time I had some time on my hands to dedicate to Thrive so I read this topic and made a quick concept.
I like tjwhales idea (I think it was his) to stick to our hexagonal system for the proteins, even if they‘re not visually represented in the cell.
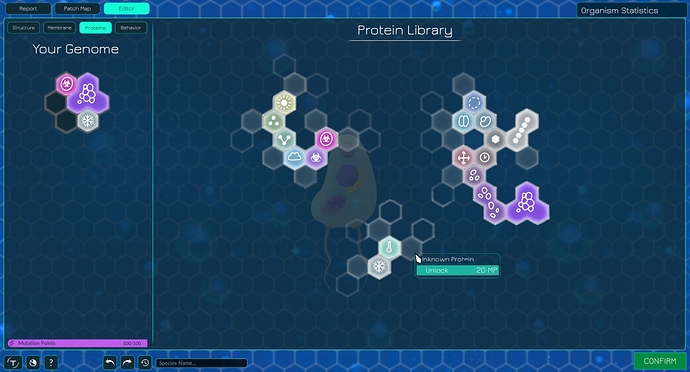
In my concept the proteins are discovered in the „protein library“. You start with three unlocked proteins (one of them enables glycolysis).You can only discover/unlock proteins that are next to a protein you already know. The player doesn‘t know exactly what he‘s unlocking next but similar proteins tend to be close to one another, so if you‘re unlocking a protein next to a toxin it‘s probably another toxin.
Speaking of toxins, toxin as well as agents in general are treated like all other enzymes/proteins. As far as I‘m aware „agent“ and „protein“ aren‘t distinct chemcial categories so I think it would be elegant to use the same system for the two.
When you unlock a protein in the library it‘s not immediatley active. First you have to assign it to a slot (or multiple slots) in your genome via drag-and-drop. While the unlocked proteins in the library represent the proteins your organism is theoretically capable of producting (the information is somewhere in your organisms DNA), the genome represents what proteins are actually being produced (the phenotype if you will). Assigned non-agent proteins are automatically active when placed in the genome. Agents first have to be further assigned to a agent gland. Every time you place an agent gland the game asks you to choose an agent from your genome that should be produced in that gland.
The capacity of your genome can be increased, most notably by adding a nucleus. Some proteins take up two or even three adjectant slots. Like this certain types of cells can be stopped from having certain kinds of proteins without the game saying „No, you can‘t use this protein“. The protein is simply too complex for the microbe and therefore doesn‘t fit in the microbes genome. Bacteria for example could only have four slots in a row in their genome. Therefore they couldn‘t use colony-building proteins as those occupy three slots in a triangle form (see image above).
First unlocking a protein in the library and then placing it in the genome (and then assigning it to a gland when it‘s an agent) might seem overly complicated, but I think the simplicity of combining the agent and the protein system as well as the interesting fitting-shapes-into-the-genome mechanics might be worth it.