Since we’re all working on some heftier other features all over the place, from sound to graphics to programming, it’s nice sometimes to have a fun and light feature to work on to destress. This is one I’ve been working on on the side, initially as a fun thought experiment but now with more purpose.
Current Naming System
Species names are generated randomly right now, with the only rules being that they must be constructed from parts that make them latin or greek sounding, and that they must be comprised of two words. The first word has a chance to not change during species splitting, to represent species that are part of the same genus.
However I’ll be honest I have a really tough time keeping up with all the species names. For example, the Patch Report before every editor is such a cool tool, but I never recognize 95% of the species names on the report, which is a shame because it ruins the value of the report for me.
For example, here are the first five names generated when I just opened the game a moment ago:
- Usian vupimpiun
- Onasmia pator
- Nora thulster (sounds like a name lol)
- Ekituian uritudata
- Mamomion occocopien
They do the job of sounding Greco-Latin-esque. But I think we could do even better.
Pitch
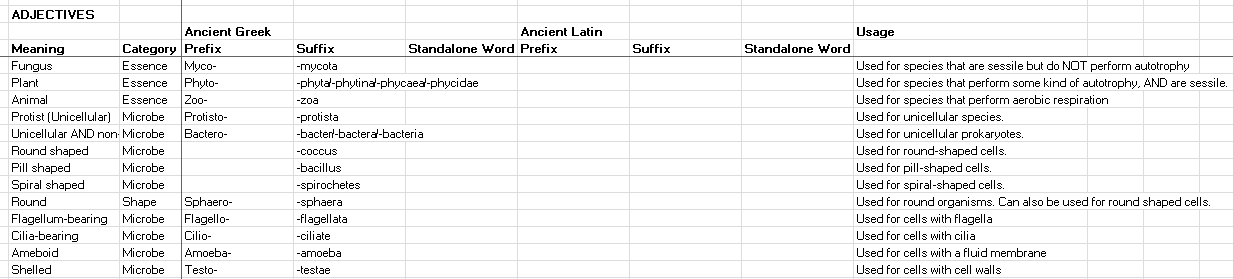
I think the Species Naming system has the potential to actually give the player a lot of hints as to the nature and relatedness of species, if instead of using Latin and Greek SOUNDING words, it actually used actual Latin and Greek prefixes/suffixes.
Here are five example names using an accurate Latin and Greek naming system:
- Euprotocyta pangonia, meaning “Pangonian true early cells”. The clade was originally named Protocyta as it was the first lineage of cells on this planet. The clade was then renamed to Euprotocyta when the Neoprotocyta clade split off from it. The species was named “pangonia” as this species originated from the Pangonian Sea.
- Euprotocyta fecundus, meaning “fertile true early cells”. This was the second species to evolve and the first to split off of the LUCA. The species was named “fecundus” because it evolved to double its rate of reproduction. However, because it didn’t evolve a “significant mutation”, it was classified as remaining within the same clade.
- Sulfurobacter polyphagus, meaning “gluttonous sulfur bacteria”. This species was the third to evolve, and the first to evolve a “significant mutation” meaning that it formed a new clade as well. The clade was named “Sulfurobacter” because the species was the first to evolve sulfur respiration (it’s defining/significant mutation). The species was named “polyphagus” because it evolved to have very large vacuoles.
- Neoprotocyta viator, meaning “Travelling new early cell”. This species evolved a chemoreceptor, which was defined as a “significant mutation” and so a new clade was formed for it. Every time a new clade needs naming, the game has a chance to not create a brand new name, but to simply modify the old clade name, to show the parentage of the new clade. In this case, it was randomly decided that this would be so, and so the new clade was named “Neoprotocyta” and the old clade was renamed to “Euprotocyta”. The species name was chosen to be “viator” since the species evolved a faster speed.
- Flagellata darwina, meaning “Darwin’s flagellates”. This species diverged from the Neoprotocyta by evolving a flagellum. The flagellum was identified as a significant mutation, and so a new clade was formed. Since this was the first species to evolve a flagellum, this clade was named “Flagellata”. The name “darwina” was assigned randomly as a species name (sometimes a random species name will be chosen if there is no meaning-based species name available).
Accurate prefixes and suffixes would accomplish the following:
- They are not gibberish. As humans we’re extraordinary at subconsciously noticing patterns, even if we don’t explicitly notice them. As such we’d immediately recognize that these are actual latin and greek suffixes, even if we’re not latin or ancient greek speakers.
- Because there is already so much latin and greek in English and many European languages as well, we will be able to intuit the meaning of many of the names. For example, if a clade was named “Protocyta”, some of you may immediately recognize that means “Early cell”, and thus you could deduce that that clade is probably the first or one of the first lineages of life.
- At a glance you will be able to tell the relationships between some clades. For example you know that the Euprotocyta and the Neoprotocyta are related.
- At a glance you will be able to tell the defining mutation of a clade. For example, you can look at the Flagellata clade and deduce they were the first lineage to evolve flagella.
- Very modular. This leaves open the possibility to create different naming system scripts we could load into the game, if we ever wanted to allow for future mods like a Sanskrit naming system, Arabic naming system, German naming system, etc.
One big question is how to define “significant mutations” to know whether to form a new clade. I have some thoughts on how to accomplish this.
Another point is that to construct such a system we’d need a big list of greek and latin prefixes and suffixes to draw from, and a robust logic system to know how to construct them into names. Luckily, I have already started on both and would be happy to carry this on in the background as a long-term project to work on until it’s finally ready. Here is a little snippet of the naming appendix I’ve been slowly filling out: