Solid-to-Gaseous Sulfur Cycle:
I agree that it would be great to see an expansion of sulfur gameplay and creating a sulfur cycle. For this, I propose that we add solid sulfur compounds to the game. These will never appear as clouds, and only as solid chunks. Cells can convert the solid chunks to H2S or covert H2S into solid chunks. This can allow more variety of food sources without having more overlapping clouds. The current reaction of hydrogen sulfide chemo-synthesis would be amended to include elemental sulfur as a product:
HydrogenSulfide + CarbonDioxide → Glucose + Sulfur
Step 1:
H2S chemosynthesis will produce tiny grains of elemental sulfur which build up in the producing cell. This can be observed in real life, as “chemosynthetic bacteria such as purple sulfur bacteria have yellow globules of sulfur visible in their cytoplasm” (Hydrogen sulfide chemosynthesis - Wikipedia). Sulfur grains build up in a cell that produces them, increasing in number or size until they reach a certain total mass, and are then expelled, becoming a single floating chunk of sulfur. The size of the chunk could depend on the storage capacity of the cell. It would have an impact on frame rates if these chunks were too small and numerous, and they would be hard to find. So, the cell filling up its storage space might be the best way to determine when to release it. It would be an automatic process.
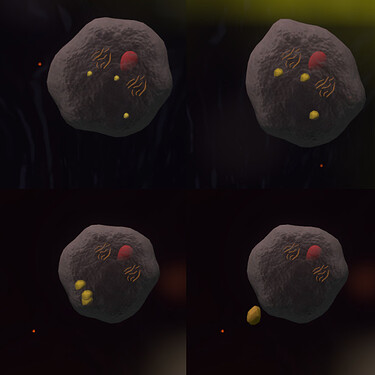
How the sulfur grains inside cells would be displayed is something others would need to weigh in on. They could be displayed as spherical meshes inside the cell, which would need to be low-poly and small enough that they don’t get confused with the organelles, it would be a smaller version of the smallest sulfur chunk currently in the game. Extra code would be needed for the cell to make them appear and grow. Another way to display them would be via the membrane shader, depicting them as yellow dots or bubbles. They would still need extra coding for the cell, but through changing the value of a single attribute of the membrane, and the shader would handle the rest. When expelling the sulfur grains, the mesh option could use a version of existing code for expelling things from the cell, and the shader option could use maths to achieve a similar-looking thing.
This is a mock-up of how they could look:
As for the nature of the floating chunks themselves, the environmental chunks would be mostly sulfate rocks of different sizes (I added alunite - Alunite - Wikipedia - in my recent PR), with some small chunks of elemental sulfur. These would exist alongside the elemental sulfur grains expelled from cells. Large Sulfur chunks could potentially release a very small amount of H2S (as they do now), and very small chunks could dissolve into H2S. Or, they could both be inert unless you have the parts to break them down. It would probably be best for the grains released from cells to dissolve, to prevent them getting too numerous.
Step 2:
For the second part of the cycle, we would also need a process to convert solid sulfur chunks to hydrogen sulfide. A new part would be added to digest solid sulfur, converting it to H2S. This could possibly be a modified version of the existing Chemosynthetic Proteins. It would work with Sulfur grains and chunks just like Rusticyanin (with siderophores) works on iron chunks - allowing storage and digestion of the chunks, and producing an agent to release small chunks from sulfur rocks. The difference would be that it wouldn’t allow digestion of any H2S that might be released from the chunks (you’d need the regular chemosynthesising protein for that).
Here’s a reaction for sulfate reduction:
SO3 → H2S + 3 H2O (Sulfite reductase - Wikipedia)
And here’s a reaction for elemental sulfur reduction:
S + H2 → H2S (Sulfur-reducing bacteria - Wikipedia, Sulfur reductase | SpringerLink)
Both of these could be represented in game as simply:
Sulfur → HydrogenSulfide
The new part could be called ‘Sulfur Reductase’, which is a real protein that performs this process with elemental sulfur or polysulfide (Sulfur reductase | SpringerLink). There would be an inbuilt assumption that we can treat elemental sulfur and sulfate the same, which requires a bit of hand-waving. Many IRL sulfate bacteria can do some processes involving both, but “although they thrive in the same habitats, sulfur reducing bacteria are incapable of sulfate reduction” (Sulfur-reducing bacteria - Wikipedia). This means that microbes can either convert elemental sulfur to H2S (S → H2S) or convert sulfate to H2S (SO4 → H2S). They can’t do both of these, although for our purposes, the two are treated the same anyway. We could refer to the solid compound simply as Sulfur (or S). It would not be necessary to display it in the compounds window, as it would show up on the cell itself.
There is a problem in this system of potential infinite recursion of the chemical processes if a cell has parts for feeding on both S and H2S - meaning that once a cell gets some sulfur, it can live on this indefinitely. There are some ways to mitigate this:
* The reactions could have less than perfect efficiency, with some sulfur being lost. This would prevent infinite re-use, and making it even less efficient would reduce the maximum lifetime of the chemicals but also make the process less desirable as a fuel source in the first place.
* We could alternatively use the Sulfate reduction reaction SO4 + ATP → H2S + 3 H2O (Sulfur cycle - Wikipedia) as a base, meaning that cells would be using energy to produce H2S. This could make it more of an investment, using extra energy when you have it to make more fuel for later.
* When the cell is producing Sulfur, this could not be counted towards usable Sulfur. The cell would have to wait until it is expelled, then swallow it again, when it would count as Ingested Matter. This wouldn’t prevent infinite recursion, but would make it more awkward to achieve.
* We could force cells to only use one pathway or the other. This would prevent any recursion for a single species, but would take a lot more work to implement.